Unravelling the means to an end: RNA polymerase II transcription termination
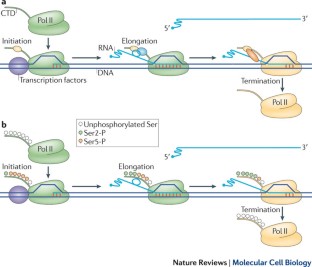
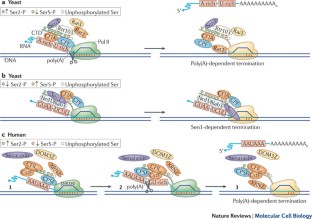
The pervasiveness of RNA synthesis in eukaryotes is largely the result of RNA polymerase II (Pol II)-mediated transcription, and termination of its activity is necessary to partition the genome and maintain the proper expression of neighbouring genes. Despite its ever-increasing biological significance, transcription termination remains one of the least understood processes in gene expression. However, recent mechanistic studies have revealed a striking convergence among several overlapping models of termination, including the poly(A)- and Sen1-dependent pathways, as well as new insights into the specificity of Pol II termination among its diverse gene targets. Broader knowledge of the role of Pol II carboxy-terminal domain phosphorylation in promoting alternative mechanisms of termination has also been gained.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
206,07 € per year
only 17,17 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Causes and consequences of RNA polymerase II stalling during transcript elongation
Article 18 November 2020

Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases
Article 03 May 2022

Structural basis for intrinsic transcription termination
Article 11 January 2023
References
- Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet.34, 77–137 (2000). CASPubMedGoogle Scholar
- Espinosa, J. M. The meaning of pausing. Mol. Cell40, 507–508 (2010). CASPubMedGoogle Scholar
- Nechaev, S. & Adelman, K. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta1809, 34–45 (2011). CASPubMedGoogle Scholar
- Rosonina, E., Kaneko, S. & Manley, J. L. Terminating the transcript: breaking up is hard to do. Genes Dev.20, 1050–1056 (2006). CASPubMedGoogle Scholar
- Gilmour, D. S. & Fan, R. Derailing the locomotive: transcription termination. J. Biol. Chem.283, 661–664 (2008). CASPubMedGoogle Scholar
- Richard, P. & Manley, J. L. Transcription termination by nuclear RNA polymerases. Genes Dev.23, 1247–1269 (2009). CASPubMedPubMed CentralGoogle Scholar
- Merino, E. & Yanofsky, C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet.21, 260–264 (2005). CASPubMedGoogle Scholar
- Naville, M. & Gautheret, D. Transcription attenuation in bacteria: theme and variations. Brief Funct. Genomics9, 178–189 (2010). CASPubMedGoogle Scholar
- Dichtl, B. Transcriptional ShortCUTs. Mol. Cell31, 617–618 (2008). CASPubMedGoogle Scholar
- Kim, K. Y. & Levin, D. E. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell144, 745–756 (2011). CASPubMedPubMed CentralGoogle Scholar
- Jacquier, A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nature Rev. Genet.10, 833–844 (2009). CASPubMedGoogle Scholar
- Shearwin, K. E., Callen, B. P. & Egan, J. B. Transcriptional interference — a crash course. Trends Genet.21, 339–345 (2005). CASPubMedPubMed CentralGoogle Scholar
- West, S. & Proudfoot, N. J. Transcriptional termination enhances protein expression in human cells. Mol. Cell33, 354–364 (2009). CASPubMedPubMed CentralGoogle Scholar
- Mapendano, C. K., Lykke-Andersen, S., Kjems, J., Bertrand, E. & Jensen, T. H. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol. Cell40, 410–422 (2010). CASPubMedGoogle Scholar
- Ardehali, M. B. & Lis, J. T. Tracking rates of transcription and splicing in vivo. Nature Struct. Mol. Biol.16, 1123–1124 (2009). CASGoogle Scholar
- Kireeva, M. L., Komissarova, N., Waugh, D. S. & Kashlev, M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem.275, 6530–6536 (2000). CASPubMedGoogle Scholar
- Komissarova, N., Becker, J., Solter, S., Kireeva, M. & Kashlev, M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell10, 1151–1162 (2002). References 16 and 17 implicate the RNA–DNA hybrid of the transcription elongation complex as a primary target of Pol II and bacterial RNA polymerase termination mechanisms.CASPubMedGoogle Scholar
- Lykke-Andersen, S. & Jensen, T. H. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie89, 1177–1182 (2007). CASPubMedGoogle Scholar
- Rondon, A. G., Mischo, H. E. & Proudfoot, N. J. Terminating transcription in yeast: whether to be a 'nerd' or a 'rat'. Nature Struct. Mol. Biol.15, 775–776 (2008). CASGoogle Scholar
- Logan, J., Falck-Pedersen, E., Darnell, J. E. Jr & Shenk, T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl Acad. Sci. USA84, 8306–8310 (1987). CASPubMedPubMed CentralGoogle Scholar
- Whitelaw, E. & Proudfoot, N. α-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α 2 globin gene. EMBO J.5, 2915–2922 (1986). CASPubMedPubMed CentralGoogle Scholar
- Edwalds-Gilbert, G., Prescott, J. & Falck-Pedersen, E. 3′ RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes. Mol. Cell. Biol.13, 3472–3480 (1993). CASPubMedPubMed CentralGoogle Scholar
- Plant, K. E., Dye, M. J., Lafaille, C. & Proudfoot, N. J. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human γ-globin gene. Mol. Cell. Biol.25, 3276–3285 (2005). CASPubMedPubMed CentralGoogle Scholar
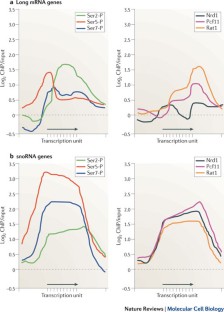
- Kim, H. et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nature Struct. Mol. Biol.17, 1279–1286 (2010). Provides genome-wide analysis of the dynamics of Pol II CTD phosphorylation and the recruitment of termination factors Pcf11, Nrd1 and Rat1.CASGoogle Scholar
- Mandel, C. R., Bai, Y. & Tong, L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci.65, 1099–1122 (2008). CASPubMedPubMed CentralGoogle Scholar
- Millevoi, S. & Vagner, S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res.38, 2757–2774 (2009). PubMedPubMed CentralGoogle Scholar
- Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell33, 365–376 (2009). CASPubMedPubMed CentralGoogle Scholar
- Egloff, S. & Murphy, S. Cracking the RNA polymerase II CTD code. Trends Genet.24, 280–288 (2008). CASPubMedGoogle Scholar
- Buratowski, S. Progression through the RNA Polymerase II CTD Cycle. Mol. Cell36, 541–546 (2009). CASPubMedPubMed CentralGoogle Scholar
- Buratowski, S. The CTD code. Nature Struct. Biol.10, 679–680 (2003). CASPubMedGoogle Scholar
- Gromak, N., West, S. & Proudfoot, N. J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol.26, 3986–3996 (2006). Shows that a pause sequence promotes poly(A)-dependent terminationin vivo, and the efficiency of termination is influenced by the strength of the poly(A) site and its proximity to the pause site.CASPubMedPubMed CentralGoogle Scholar
- Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D. L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature Struct. Mol. Biol.15, 71–78 (2008). CASGoogle Scholar
- Park, N. J., Tsao, D. C. & Martinson, H. G. The two steps of poly(A)-dependent termination, pausing and release, can be uncoupled by truncation of the RNA polymerase II carboxyl-terminal repeat domain. Mol. Cell. Biol.24, 4092–4103 (2004). CASPubMedPubMed CentralGoogle Scholar
- Nag, A., Narsinh, K. & Martinson, H. G. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nature Struct. Mol. Biol.14, 662–669 (2007). CASGoogle Scholar
- Kazerouninia, A., Ngo, B. & Martinson, H. G. Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA16, 197–210 (2010). References 33–35 from the Martinson laboratory show that poly(A)-dependent termination of Pol II can be separated into two steps, pausing and release, which depend on interactions of the cleavage and polyadenylation machinery with the body of Pol II and the Pol II CTD.CASPubMedPubMed CentralGoogle Scholar
- Alexander, R. D., Innocente, S. A., Barrass, J. D. & Beggs, J. D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell40, 582–593 (2010). CASPubMedPubMed CentralGoogle Scholar
- Carrillo Oesterreich, F., Preibisch, S. & Neugebauer, K. M. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell40, 571–581 (2010). CASPubMedGoogle Scholar
- Kim, M. et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature432, 517–522 (2004). CASPubMedGoogle Scholar
- West, S., Gromak, N. & Proudfoot, N. J. Human 5′–3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature432, 522–525 (2004). References 38 and 39 demonstrate that the 5′–3′ exoribonuclease Rat1 (XRN2 in mammals) is important for poly(A)-dependent termination and strengthen support for the torpedo model.CASPubMedGoogle Scholar
- Luo, W., Johnson, A. W. & Bentley, D. L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev.20, 954–965 (2006). Reveals that in addition to its exoribonuclease activity, Rat1 helps recruit mRNA 3′-end-processing factors. The authors propose a model for the termination mechanism that incorporates both allosteric and torpedo components.CASPubMedPubMed CentralGoogle Scholar
- Lunde, B. M. et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nature Struct. Mol. Biol.17, 1195–1201 (2010). CASGoogle Scholar
- Teixeira, A. et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature432, 526–530 (2004). CASPubMedGoogle Scholar
- Ghazal, G. et al. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol. Cell36, 99–109 (2009). CASPubMedGoogle Scholar
- Rondón, A., Mischo, H., Kawauchi, J. & Proudfoot, N. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae.Mol. Cell36, 88–98 (2009). PubMedPubMed CentralGoogle Scholar
- Nabavi, S. & Nazar, R. N. Pac1 endonuclease and Dhp1p 5′–3′ exonuclease are required for U3 snoRNA termination in Schizosaccharomyces pombe. FEBS Lett.584, 3436–3441 (2010). CASPubMedGoogle Scholar
- Connelly, S. & Manley, J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev.2, 440–452 (1988). CASPubMedGoogle Scholar
- Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell136, 763–776 (2009). CASPubMedGoogle Scholar
- Kim, H. D., Choe, J. & Seo, Y. S. The sen1 + gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry38, 14697–14710 (1999). CASPubMedGoogle Scholar
- Steinmetz, E. J. & Brow, D. A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol.16, 6993–7003 (1996). CASPubMedPubMed CentralGoogle Scholar
- Steinmetz, E. J., Conrad, N. K., Brow, D. A. & Corden, J. L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature413, 327–331 (2001). CASPubMedGoogle Scholar
- Steinmetz, E. J. et al. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell24, 735–746 (2006). This paper establishes Sen1 as a general transcription termination factor in yeast for most snRNAs and snoRNAs and some short mRNA transcripts.CASPubMedGoogle Scholar
- Ursic, D., Chinchilla, K., Finkel, J. S. & Culbertson, M. R. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res.32, 2441–2452 (2004). CASPubMedPubMed CentralGoogle Scholar
- Finkel, J. S., Chinchilla, K., Ursic, D. & Culbertson, M. R. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics184, 107–118 (2010). CASPubMedPubMed CentralGoogle Scholar
- Mischo, H. E. et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell41, 21–32 (2011). CASPubMedPubMed CentralGoogle Scholar
- Arigo, J. T., Eyler, D. E., Carroll, K. L. & Corden, J. L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell23, 841–851 (2006). CASPubMedGoogle Scholar
- Thiebaut, M., Kisseleva-Romanova, E., Rougemaille, M., Boulay, J. & Libri, D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the Nrd1-Nab3 pathway in genome surveillance. Mol. Cell23, 853–864 (2006). CASPubMedGoogle Scholar
- Banerjee, S., Chalissery, J., Bandey, I. & Sen, R. Rho-dependent transcription termination: more questions than answers. J. Microbiol.44, 11–22 (2006). CASPubMedPubMed CentralGoogle Scholar
- Kawauchi, J., Mischo, H., Braglia, P., Rondon, A. & Proudfoot, N. J. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev.22, 1082–1092 (2008). CASPubMedPubMed CentralGoogle Scholar
- Banerjee, A., Sammarco, M. C., Ditch, S., Wang, J. & Grabczyk, E. A novel tandem reporter quantifies RNA polymerase II termination in mammalian cells. PLoS ONE4, e6193 (2009). PubMedPubMed CentralGoogle Scholar
- Suraweera, A. et al. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet.18, 3384–3396 (2009). CASPubMedGoogle Scholar
- Matera, A. G., Terns, R. M. & Terns, M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nature Rev. Mol. Cell Biol.8, 209–220 (2007). CASGoogle Scholar
- Egloff, S. et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science318, 1777–1779 (2007). This study reveals that Ser7-P Pol II CTD helps recruit the integrator complex, which is required for human snRNA 3′-end processing.CASPubMedPubMed CentralGoogle Scholar
- Ezzeddine, N. et al. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′ end formation. Mol. Cell. Biol.31, 328–341 (2011). CASPubMedGoogle Scholar
- Baillat, D. et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell123, 265–276 (2005). CASPubMedGoogle Scholar
- Dominski, Z., Yang, X.-C., Purdy, M., Wagner, E. J. & Marzluff, W. F. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol.25, 1489–1500 (2005). CASPubMedPubMed CentralGoogle Scholar
- Egloff, S., Al-Rawaf, H., O'Reilly, D. & Murphy, S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and β-actin genes. Mol. Cell. Biol.29, 4002–4013 (2009). CASPubMedPubMed CentralGoogle Scholar
- Ballarino, M. et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol.29, 5632–5638 (2009). CASPubMedPubMed CentralGoogle Scholar
- Carninci, P. Molecular biology: the long and short of RNAs. Nature457, 974–975 (2009). CASPubMedGoogle Scholar
- Dengl, S. & Cramer, P. Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro. J. Biol. Chem.284, 21270–21279 (2009). CASPubMedPubMed CentralGoogle Scholar
- Saeki, H. & Svejstrup, J. Q. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell35, 191–205 (2009). CASPubMedPubMed CentralGoogle Scholar
- Xiang, S. et al. Structure and function of the 5′–3′ exoribonuclease Rat1 and its activating partner Rail. Nature458, 784–788 (2009). CASPubMedPubMed CentralGoogle Scholar
- Chang, J. H. & Xiang, S. Structural and biochemical studies of the 5′–3′ exoribonuclease Xrnl. Nature Cell Biol.18, 270–276 (2011). CASGoogle Scholar
- Epshtein, V., Dutta, D., Wade, J. & Nudler, E. An allosteric mechanism of Rho-dependent transcription termination. Nature463, 245–249 (2010). Demonstrates that the Rho termination factor associates directly withE. coliRNA polymerase and promotes termination by inducing an allosteric rearrangement of the RNA polymerase active site. Transduction of the termination signal is dependent on the lid and trigger loop domains of the RNA polymerase β′-subunit.CASPubMedPubMed CentralGoogle Scholar
- Lang, W. H., Platt, T. & Reeder, R. H. Escherichia coli Rho factor induces release of yeast RNA polymerase II but not polymerase I or III. Proc. Natl Acad. Sci. USA95, 4900–4905 (1998). CASPubMedPubMed CentralGoogle Scholar
- Schmidt, M. C. & Chamberlin, M. J. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J. Mol. Biol.195, 809–818 (1987). CASPubMedGoogle Scholar
- Sullivan, S. L. & Gottesman, M. E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell68, 989–994 (1992). CASPubMedGoogle Scholar
- Mason, S. W., Li, J. & Greenblatt, J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage λ. J. Biol. Chem.267, 19418–19426 (1992). CASPubMedGoogle Scholar
- Torres, M., Condon, C., Balada, J. M., Squires, C. & Squires, C. L. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J.20, 3811–3820 (2001). CASPubMedPubMed CentralGoogle Scholar
- Shankar, S., Hatoum, A. & Roberts, J. W. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol. Cell27, 914–927 (2007). CASPubMedPubMed CentralGoogle Scholar
- Ha, K. S., Toulokhonov, I., Vassylyev, D. G. & Landick, R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol.401, 708–725 (2010). Findings from this study show that interaction of the NusA termination factor with regions of bacterial RNA polymerase near the RNA exit channel (such as the β-flap and β′-dock) stimulates pausing and release.CASPubMedPubMed CentralGoogle Scholar
- Toulokhonov, I., Artsimovitch, I. & Landick, R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science292, 730–733 (2001). CASPubMedGoogle Scholar
- Deighan, P., Diez, C. M., Leibman, M., Hochschild, A. & Nickels, B. E. The bacteriophage λQ antiterminator protein contacts the β-flap domain of RNA polymerase. Proc. Natl Acad. Sci. USA105, 15305–15310 (2008). CASPubMedPubMed CentralGoogle Scholar
- Mooney, R. A., Schweimer, K., Rösch, P., Gottesman, M. & Landick, R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol.391, 341–358 (2009). Reveals how interactions with two separate protein domains of NusG contribute to its RNA polymerase termination and antitermination activities. The NusG NTD contacts the RNA polymerase β′-clamp helices while its CTD binds Rho or other transcriptional regulators.CASPubMedPubMed CentralGoogle Scholar
- Nickels, B. E. Genetic assays to define and characterize protein–protein interactions involved in gene regulation. Methods47, 53–62 (2009). CASPubMedGoogle Scholar
- Belogurov, G. A., Mooney, R. A., Svetlov, V., Landick, R. & Artsimovitch, I. Functional specialization of transcription elongation factors. EMBO J.28, 112–122 (2009). CASPubMedGoogle Scholar
- Belogurov, G. A., Sevostyanova, A., Svetlov, V. & Artsimovitch, I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol. Microbiol.76, 286–301 (2010). CASPubMedPubMed CentralGoogle Scholar
- Komarnitsky, P., Cho, E. J. & Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev.14, 2452–2460 (2000). Shows that dynamic phosphorylation of the Pol II CTD contributes to differential recruitment of mRNA processing factors, a discovery that forms the basis of the CTD code hypothesis.CASPubMedPubMed CentralGoogle Scholar
- Licatalosi, D. D. et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell9, 1101–1111 (2002). CASPubMedGoogle Scholar
- Ahn, S. H., Kim, M. & Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell13, 67–76 (2004). CASPubMedGoogle Scholar
- Chapman, R. D. et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science318, 1780–1782 (2007). Identifies dynamic phosphorylation of the Pol II CTD at Ser7as a new component of the CTD code.CASPubMedGoogle Scholar
- Guo, J., Garrett, M., Micklem, G. & Brogna, S. Poly(A) signals located near the 5′ end of genes are silenced by a general mechanism that prevents premature 3′-end processing. Mol. Cell. Biol.31, 639–651 (2011). CASPubMedGoogle Scholar
- Gudipati, R. K., Villa, T., Boulay, J. & Libri, D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nature Struct. Mol. Biol.15, 786–794 (2008). Shows that the selection of the poly(A)-dependent versus Sen1-dependent termination pathway is influenced by the phosphorylation status of the Pol II CTD, which is in turn influenced by the distance that Pol II travels from the transcription start site.CASGoogle Scholar
- Jenks, M. H., O'Rourke, T. W. & Reines, D. Properties of an intergenic terminator and start site switch that regulate IMD2 transcription in yeast. Mol. Cell. Biol.28, 3883–3893 (2008). CASPubMedPubMed CentralGoogle Scholar
- Vasiljeva, L., Kim, M., Mutschler, H., Buratowski, S. & Meinhart, A. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nature Struct. Mol. Biol.15, 795–804 (2008). CASGoogle Scholar
- Mayer, A. et al. Uniform transitions of the general RNA polymerase II transcription complex. Nature Struct. Mol. Biol.17, 1272–1278 (2010). CASGoogle Scholar
- Tietjen, J. R. et al. Chemical-genomic dissection of the CTD code. Nature Struct. Mol. Biol.17, 1154–1161 (2010). References 95 and 96 provide genome-wide analyses of the dynamics of Pol II CTD-phosphorylation and identify gene-specific patterns of CTD marks or transcription factor recruitment, respectively.CASGoogle Scholar
- Kim, M. et al. Distinct pathways for snoRNA and mRNA termination. Mol. Cell24, 723–734 (2006). Reveals that many poly(A)-dependent and Sen1-dependent termination factors can be localized to both mRNA and snoRNA-encoding genes, but their requirements for termination are gene-specific.CASPubMedGoogle Scholar
- Egloff, S. et al. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem.285, 20564–20569 (2010). CASPubMedPubMed CentralGoogle Scholar
- Singh, N. et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell36, 255–266 (2009). CASPubMedPubMed CentralGoogle Scholar
- Werner-Allen, J. W. et al. cis-proline-mediated Ser(P) 5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem.286, 5717–5726 (2011). CASPubMedGoogle Scholar
- Selth, L. A., Sigurdsson, S. & Svejstrup, J. Q. Transcript Elongation by RNA Polymerase II. Annu. Rev. Biochem.79, 271–293 (2010). CASPubMedGoogle Scholar
- Kim, M., Ahn, S. H., Krogan, N. J., Greenblatt, J. F. & Buratowski, S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J.23, 354–364 (2004). CASPubMedPubMed CentralGoogle Scholar
- Jaehning, J. A. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta1799, 379–388 (2010). CASPubMedPubMed CentralGoogle Scholar
- Alén, C. et al. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell10, 1441–1452 (2002). PubMedGoogle Scholar
- Wood, A. J. et al. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev.22, 1141–1146 (2008). CASPubMedPubMed CentralGoogle Scholar
- Spies, N., Nielsen, C. B., Padgett, R. A. & Burge, C. B. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell36, 245–254 (2009). CASPubMedPubMed CentralGoogle Scholar
- Alló, M. & Kornblihtt, A. R. Gene silencing: small RNAs control RNA polymerase II elongation. Curr. Biol.20, R704–R707 (2010). PubMedGoogle Scholar
- Fan, X. et al. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc. Natl Acad. Sci. USA107, 17945–17950 (2010). CASPubMedPubMed CentralGoogle Scholar
- Mayr, C. & Bartel, D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell138, 673–684 (2009). CASPubMedPubMed CentralGoogle Scholar
- Calvo, O. & Manley, J. L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev.17, 1321–1327 (2003). CASPubMedGoogle Scholar
- Venters, B. J. & Pugh, B. F. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol.44, 117–141 (2009). CASPubMedPubMed CentralGoogle Scholar
- Hampsey, M., Singh, B. N., Ansari, A., Lainé, J.-P. & Krishnamurthy, S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv. Enzyme Regul. 29 Oct 2010 (doi:10.1016/j.advenzreg.2010.10.001). CASPubMedGoogle Scholar
- El Kaderi, B., Medler, S., Raghunayakula, S. & Ansari, A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem.284, 25015–25025 (2009). CASPubMedPubMed CentralGoogle Scholar
- Wang, Y., Fairley, J. A. & Roberts, S. G. E. Phosphorylation of TFIIB links transcription initiation and termination. Curr. Biol.20, 548–553 (2010). CASPubMedPubMed CentralGoogle Scholar
- Moore, M. J. & Proudfoot, N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell136, 688–700 (2009). CASPubMedGoogle Scholar
- Lykke-Andersen, S., Mapendano, C. K. & Jensen, T. H. An ending is a new beginning: transcription termination supports re-initiation. Cell Cycle10, 863–865 (2011). CASPubMedGoogle Scholar
- Glover-Cutter, K. et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol.29, 5455–5464 (2009). CASPubMedPubMed CentralGoogle Scholar
- Jimeno-González, S., Haaning, L. L., Malagon, F. & Jensen, T. H. The yeast 5′-3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol. Cell37, 580–587 (2010). PubMedGoogle Scholar
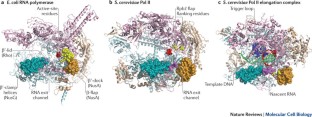
- Opalka, N. et al. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol.8, e1000483 (2010). PubMedPubMed CentralGoogle Scholar
- Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science292, 1863–1876 (2001). CASPubMedGoogle Scholar
- Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell16, 955–965 (2004). CASPubMedGoogle Scholar
- Birse, C. E., Minvielle-Sebastia, L., Lee, B. A., Keller, W. & Proudfoot, N. J. Coupling termination of transcription to messenger RNA maturation in yeast. Science280, 298–301 (1998). CASPubMedGoogle Scholar
- Sadowski, M., Dichtl, B., Hübner, W. & Keller, W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J.22, 2167–2177 (2003). CASPubMedPubMed CentralGoogle Scholar
- Zhang, Z., Fu, J. & Gilmour, D. S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev.19, 1572–1580 (2005). CASPubMedPubMed CentralGoogle Scholar
- Dichtl, B. et al. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J.21, 4125–4135 (2002). CASPubMedPubMed CentralGoogle Scholar
- Garas, M., Dichtl, B. & Keller, W. The role of the putative 3′ end processing endonuclease Ysh1p in mRNA and snoRNA synthesis. RNA14, 2671–2684 (2008). CASPubMedPubMed CentralGoogle Scholar
- Nedea, E. et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem.278, 33000–33010 (2003). CASPubMedGoogle Scholar
- Nedea, E. et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell29, 577–587 (2008). CASPubMedGoogle Scholar
- Ghazy, M. A., He, X., Singh, B. N., Hampsey, M. & Moore, C. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol. Cell. Biol.29, 2296–2307 (2009). CASPubMedPubMed CentralGoogle Scholar
- Ganem, C. et al. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J.22, 1588–1598 (2003). CASPubMedPubMed CentralGoogle Scholar
- Steinmetz, E. J. & Brow, D. A. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol.23, 6339–6349 (2003). CASPubMedPubMed CentralGoogle Scholar
- Xue, Y. et al. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol.20, 4006–4015 (2000). CASPubMedPubMed CentralGoogle Scholar
- Kaneko, S., Rozenblatt-Rosen, O., Meyerson, M. & Manley, J. L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev.21, 1779–1789 (2007). CASPubMedPubMed CentralGoogle Scholar
- Arigo, J. T., Carroll, K. L., Ames, J. M. & Corden, J. L. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell21, 641–651 (2006). CASPubMedGoogle Scholar
- Barillà, D., Lee, B. A. & Proudfoot, N. J. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA98, 445–450 (2001). PubMedPubMed CentralGoogle Scholar
- McCracken, S. et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature385, 357–361 (1997). CASPubMedGoogle Scholar
- Steinmetz, E. J., Ng, S. B., Cloute, J. P. & Brow, D. A. cis- and trans-acting determinants of transcription termination by yeast RNA polymerase II. Mol. Cell. Biol.26, 2688–2696 (2006). CASPubMedPubMed CentralGoogle Scholar
- Mueller, C. L., Porter, S. E., Hoffman, M. G. & Jaehning, J. A. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell14, 447–456 (2004). CASPubMedGoogle Scholar
- Sheldon, K. E., Mauger, D. M. & Arndt, K. M. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell20, 225–236 (2005). CASPubMedPubMed CentralGoogle Scholar
Acknowledgements
Research in the laboratory of C.M. is supported by grants from the US National Institutes of Health National Institute of General Medical Sciences: award numbers K12GM074869 (J.N.K.), R01GM041752 (C.M.) and R01GM068887 (C.M.). We thank G. Meinke for assistance in preparing PyMOL images.








